K-means¶
K-means is a classic method for clustering or vector quantization. The K-means algorithms produces a fixed number of clusters, each associated with a center (also known as a prototype), and each sample belongs to a cluster with the nearest center.
From a mathematical standpoint, K-means is a coordinate descent algorithm to solve the following optimization problem:
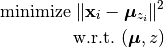
Here, 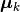 is the center of the
is the center of the k-th cluster, and 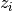 indicates the cluster for
indicates the cluster for 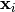 .
.
This package implements the K-means algorithm in the kmeans function:
-
kmeans(X, k; ...)¶ Performs K-means clustering over the given dataset.
Parameters: - X – The given sample matrix. Each column of
Xis a sample. - k – The number of clusters.
This function returns an instance of
KmeansResult, which is defined as follows:type KmeansResult{T<:FloatingPoint} <: ClusteringResult centers::Matrix{T} # cluster centers, size (d, k) assignments::Vector{Int} # assignments, length n costs::Vector{T} # costs of the resultant assignments, length n counts::Vector{Int} # number of samples assigned to each cluster, length k cweights::Vector{Float64} # cluster weights, length k totalcost::Float64 # total cost (i.e. objective) iterations::Int # number of elapsed iterations converged::Bool # whether the procedure converged end
One may optionally specify some of the options through keyword arguments to control the algorithm:
name description default initInitialization algorithm or initial seeds, which can be either of the following:
- a symbol indicating the name of seeding algorithm,
:rand,:kmpp, or:kmcen(see Clustering Initialization) - an integer vector of length
kthat provides the indexes of initial seeds.
:kmppmaxiterMaximum number of iterations. 100tolTolerable change of objective at convergence. 1.0e-6weightsThe weights of samples, which can be either of:
nothing: each sample has a unit weight.- a vector of length
nthat gives the sample weights.
nothingdisplayThe level of information to be displayed. (see Common Options) :none- X – The given sample matrix. Each column of
If you already have a set of initial center vectors, you may use kmeans! instead:
-
kmeans!(X, centers; ...) Performs K-means given initial centers, and updates the centers inplace.
Parameters: - X – The given sample matrix. Each column of
Xis a sample. - centers – The matrix of centers. Each column of
centersis a center vector for a cluster.
Note: The number of clusters
kis determined assize(centers, 2).Like
kmeans, this function returns an instance ofKmeansResult.This function accepts all keyword arguments listed above for
kmeans(exceptinit).- X – The given sample matrix. Each column of
Examples:
using Clustering
# make a random dataset with 1000 points
# each point is a 5-dimensional vector
X = rand(5, 1000)
# performs K-means over X, trying to group them into 20 clusters
# set maximum number of iterations to 200
# set display to :iter, so it shows progressive info at each iteration
R = kmeans(X, 20; maxiter=200, display=:iter)
# the number of resultant clusters should be 20
@assert nclusters(R) == 20
# obtain the resultant assignments
# a[i] indicates which cluster the i-th sample is assigned to
a = assignments(R)
# obtain the number of samples in each cluster
# c[k] is the number of samples assigned to the k-th cluster
c = counts(R)
# get the centers (i.e. mean vectors)
# M is a matrix of size (5, 20)
# M[:,k] is the mean vector of the k-th cluster
M = R.centers
Example with plot
using RDatasets
iris = dataset("datasets", "iris")
head(iris)
# K-means Clustering unsupervised machine learning example
using Clustering
features = permutedims(convert(Array, iris[:,1:4]), [2, 1]) # use matrix() on Julia v0.2
result = kmeans( features, 3 ) # onto 3 clusters
using Gadfly
plot(iris, x = "PetalLength", y = "PetalWidth", color = result.assignments, Geom.point)